AWS Feed
Build a medical sentence matching application using BERT and Amazon SageMaker

Determining the relevance of a sentence when compared to a specific document is essential for many different types of applications across various industries. In this post, we focus on a use case within the healthcare field to help determine the accuracy of information regarding patient health.
Frequently, during each patient visit, a new document is created with the information from the visit. This information often consists of a medical transcription that has been dictated by either the nurse or the physician. Such a document may contain a brief description statement (also known as a restatement) that explains the main details from that specific patient visit. In future visits, doctors may rely on previous visits’ restatements to quickly get an overview of the patient’s overall status. Such restatements may also be used during patient handoffs. However, this introduces the potential for errors to be made during patient handoffs to new medical teams if the restatements are difficult to understand or if they contain inadequate information (Staggers et. al. 2011). Therefore, having an accurate description of the patient’s status is important, because the cost of errors in such restatements can be high and may negatively affect the patient’s overall care (Garcia et. al. 2017).
This post walks you through how to deploy a machine learning (ML) model that aims to determine the top sentences from the document that best match the corresponding document restatement; this can be a first step to ensure the accuracy of the patient’s health records overall by determining the relevance of the restatement. We emphasize that this model determines the top ranking sentences that match the restatement; it does not generate the restatement itself.
When creating this solution, we were faced with a dual-sided challenge. Beyond the technical challenge of actually creating an AI/ML model, several surrounding components complicate actually using such models in the real world. Indeed, the actual ML code may be a very small part of the system as a whole (Sculley et al. 2015). This is especially so in complex architectures frequently deployed in the context of the healthcare and life science space.
We focused on one particular challenge: creating the ability to serve the model so that others (applications, services, or people) can use it. By serving a model, we mean to grant others the ability to pass new data to the model so they can get the predictions they need. This post provides a broad overview of the problem, the solution, and a few points to keep in mind if you plan to use a similar approach in your own use cases. A full technical write up, including a readme and a step-by-step deployment of the architecture, is available in the GitHub code repository. For more information about approaches to serving models, see Build, Train, and Deploy a Machine Learning Model With Amazon SageMaker and AWS Deep Learning Containers on Amazon ECS.
Background and use case
In the medical field (as well as other industries), documents are frequently associated with a shorter restatement text of the original document. We use the term restatement, but in fact this shorter text can be a summary, highlight, description, or other metadata about the document. For example, an after-visit clinical summary given to a patient summarizes the content of the patient visit to a physician.
For illustration purposes, the following is an example that’s unrelated to the medical industry.
Document:
On Monday morning, Joshua ate a large breakfast of bacon and eggs. He then went for a brisk walk. Finally, he returned home and sat at his desk.
Restatement:
Joshua went for a walk.
In this example, the restatement is just a rewording of the highlighted sentence in the full document. This example shows that, although the use case that we focus on in this post is specific to the medical field, you can use and modify this approach for many other text analysis applications.
Let’s now take a closer look at the use case for this post. We used data taken from MTSamples (which we downloaded from Kaggle). This data contains many different samples of transcribed medical texts. It includes documents with raw transcriptions of sample notes, as well as shorter descriptions of those notes (which we treat as restatements).
The following is an example from the MTSamples dataset.
Document:
HISTORY OF PRESENT ILLNESS: , I have seen ABC today. He is a very pleasant gentleman who is 42 years old, 344 pounds. He is 5’9″. He has a BMI of 51. He has been overweight for ten years since the age of 33, at his highest he was 358 pounds, at his lowest 260. He is pursuing surgical attempts of weight loss to feel good, get healthy, and begin to exercise again. He wants to be able to exercise and play volleyball. Physically, he is sluggish. He gets tired quickly. He does not go out often. When he loses weight he always regains it and he gains back more than he lost. His biggest weight loss is 25 pounds and it was three months before he gained it back. He did six months of not drinking alcohol and not taking in many calories. He has been on multiple commercial weight loss programs including Slim Fast for one month one year ago and Atkin’s Diet for one month two years ago.,PAST MEDICAL HISTORY: , He has difficulty climbing stairs, difficulty with airline seats, tying shoes, used to public seating, difficulty walking, high cholesterol, and high blood pressure. He has asthma and difficulty walking two blocks or going eight to ten steps. He has sleep apnea and snoring. He is a diabetic, on medication. He has joint pain, knee pain, back pain, foot and ankle pain, leg and foot swelling. He has hemorrhoids.,PAST SURGICAL HISTORY: , Includes orthopedic or knee surgery.,SOCIAL HISTORY: , He is currently single. He drinks alcohol ten to twelve drinks a week, but does not drink five days a week and then will binge drink. He smokes one and a half pack a day for 15 years, but he has recently stopped smoking for the past two weeks.,FAMILY HISTORY: , Obesity, heart disease, and diabetes. Family history is negative for hypertension and stroke.,CURRENT MEDICATIONS:, Include Diovan, Crestor, and Tricor.,MISCELLANEOUS/EATING HISTORY: ,He says a couple of friends of his have had heart attacks and have had died. He used to drink everyday, but stopped two years ago. He now only drinks on weekends. He is on his second week of Chantix, which is a medication to come off smoking completely. Eating, he eats bad food. He is single. He eats things like bacon, eggs, and cheese, cheeseburgers, fast food, eats four times a day, seven in the morning, at noon, 9 p.m., and 2 a.m. He currently weighs 344 pounds and 5’9″. His ideal body weight is 160 pounds. He is 184 pounds overweight. If he lost 70% of his excess body weight that would be 129 pounds and that would get him down to 215.,REVIEW OF SYSTEMS: , Negative for head, neck, heart, lungs, GI, GU, orthopedic, or skin. He also is positive for gout. He denies chest pain, heart attack, coronary artery disease, congestive heart failure, arrhythmia, atrial fibrillation, pacemaker, pulmonary embolism, or CVA. He denies venous insufficiency or thrombophlebitis. Denies shortness of breath, COPD, or emphysema. Denies thyroid problems, hip pain, osteoarthritis, rheumatoid arthritis, GERD, hiatal hernia, peptic ulcer disease, gallstones, infected gallbladder, pancreatitis, fatty liver, hepatitis, rectal bleeding, polyps, incontinence of stool, urinary stress incontinence, or cancer. He denies cellulitis, pseudotumor cerebri, meningitis, or encephalitis.,PHYSICAL EXAMINATION: ,He is alert and oriented x 3. Cranial nerves II-XII are intact. Neck is soft and supple. Lungs: He has positive wheezing bilaterally. Heart is regular rhythm and rate. His abdomen is soft. Extremities: He has 1+ pitting edema.,IMPRESSION/PLAN:, I have explained to him the risks and potential complications of laparoscopic gastric bypass in detail and these include bleeding, infection, deep venous thrombosis, pulmonary embolism, leakage from the gastrojejuno-anastomosis, jejunojejuno-anastomosis, and possible bowel obstruction among other potential complications. He understands. He wants to proceed with workup and evaluation for laparoscopic Roux-en-Y gastric bypass. He will need to get a letter of approval from Dr. XYZ. He will need to see a nutritionist and mental health worker. He will need an upper endoscopy by either Dr. XYZ. He will need to go to Dr. XYZ as he previously had a sleep study. We will need another sleep study. He will need H. pylori testing, thyroid function tests, LFTs, glycosylated hemoglobin, and fasting blood sugar. After this is performed, we will submit him for insurance approval.
Restatement:
Consult for laparoscopic gastric bypass.
Although the raw transcript document is quite long, only a few of the sentences actually appear to be related to the restatement “Consult for laparoscopic gastric bypass.” We highlighted two sentences within the document that you might intuitively think best match the restatement. The approach we deployed quantifies the similarities and reports the sentences in the document that best match the restatement. We did this by using a pretrained BERT language model trained specifically on clinical texts (published by Alsentzer et. al. 2019). The model itself is hosted by HuggingFace, a platform for sharing open-source natural language processing (NLP) projects. We used this model to calculate sentence-by-sentence similarities using the sentence-transform Python library.
It is important to note that in this example and in this solution, we are performing the sentence ranking without explicitly extracting and detecting the medical entities. However, many applications rely on explicitly extracting and analyzing diagnoses, medications, and other health information. For detecting medical entities such as medical conditions, medications, and other medical information in medical text, consider using Amazon Comprehend Medical, a HIPAA-eligible service built to extract medical information from unstructured medical text.
More information about this approach is available in our technical write-up.
Architecture diagram
In this section, we go over the architecture diagram for this solution at a very high level. For more details and to see the step-by-step framework, see our technical write-up.

In the model development and testing phase, we use Amazon SageMaker Studio. Studio is a powerful integrated development environment (IDE) for building, training, testing, and deploying ML models. Because we use a prebuilt model for this solution, we don’t need to use Studio’s full ability to train algorithms at scale. Instead, we use it for development and deployment purposes.
We created a Jupyter notebook that you can import into Studio. This notebook walks you through the entire development and deployment process. We start by writing the code for our model to a file. The model is then built using an NGINX/Flask framework, so that new data can be passed to it at inference time. Prior to deploying the model, we package it as a Docker container, build it using AWS CodeBuild, and push it to Amazon Elastic Container Registry (Amazon ECR). Then we deploy the model using Amazon Elastic Container Service (Amazon ECS).
The final result is a model that you can query using a simple API call. This is an important point: the ability to query models via an API capability is an essential component of designing scalable, easy-to-use interfaces. For more information, see Implementing Microservices on AWS.
After we deploy our model, we create a graphical user interface (using Streamlit) so that our model can be easily accessed through a webpage. Streamlit is an open-source library used to create front ends for ML applications. After we create our webpage, we deploy it in a similar way to how we deployed our model: we package it as a separate Docker container, build it using CodeBuild, push it to Amazon ECR, and deploy it using Amazon ECS.
By creating and deploying this webpage, we provide users with no programming experience the ability to use our model to test their own documents and restatements. The following screenshot shows what the webpage looks like.

After the user inputs their restatement and corresponding document, the top five results (the five sentences that best match the statement) are returned. If you deploy the entire solution using our original MTSamples example, the final result looks like the following screenshot.
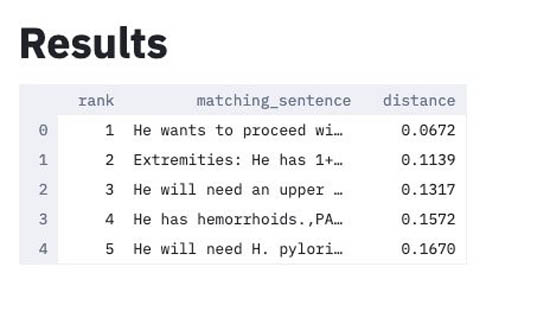
The solution reports the following results:
- The top five sentences within the document that best match the restatement.
- The similarity distance between each sentence and the restatement. A lower distance means closer similarities between that sentence and the restatement sentence.
In this example, the best matching sentence is “He wants to proceed with workup and evaluation for laparoscopic Roux-en-Y gastric bypass” with a distance of .0672. Therefore, this approach has correctly identified a sentence within the document that matches the restatement.
Limitations
Like any algorithm, this approach has some limitations. For instance, this approach is not designed to handle cases where the restatement of the document is actually high-level metadata about the document not directly related to the text of the document itself. You can solve such use cases by using Amazon Comprehend custom models. For more information, see Comprehend Custom and Building a custom classifier using Amazon Comprehend.
Another limitation in our approach is that it doesn’t explicitly handle negation (words such as “not,” “no,” and “denies”), which may change the meaning of the text. AWS services such as Amazon Comprehend and Amazon Comprehend Medical use deep learning models to handle negation.
Conclusion
In this post, we walked through the high-level steps to deploy a pre-built NLP model to analyze medical texts. If you’re interested in deploying this yourself, see our step-by-step technical write-up.
References
For more information, see the following references:
- Pretrained model – Bio_ClinicalBERT
- Sample data – Medical Transcriptions
- Original data source – MTSamples
- Basis for containers used to deploy the BERT-based model – GitHub repo
- Additional journal referenced – Why patient summaries in electronic health records do not provide the cognitive support necessary for nurses’ handoffs on medical and surgical units: Insights from interviews and observations
- Additional journal referenced – Clinical Summarization Capabilities of Commercially-available and Internally-developed Electronic Health Records
About the Authors
 Joshua Broyde is an AI/ML Specialist Solutions Architect on the Global Healthcare and Life Sciences team at Amazon Web Services. He works with customers in the healthcare and life sciences industry at all levels of the Machine Learning Lifecycle on a number of AI/ML fronts, including analyzing medical images and video, analyzing machine sensor data and performing natural language processing of medical and healthcare texts.
Joshua Broyde is an AI/ML Specialist Solutions Architect on the Global Healthcare and Life Sciences team at Amazon Web Services. He works with customers in the healthcare and life sciences industry at all levels of the Machine Learning Lifecycle on a number of AI/ML fronts, including analyzing medical images and video, analyzing machine sensor data and performing natural language processing of medical and healthcare texts.
 Claire Palmer is a Solutions Architect at Amazon Web Services. She is on the Global Account Development team, supporting healthcare and life sciences customers. Claire has a passion for driving innovation initiatives and developing solutions that are both secure and scalable. She is based out of Seattle, Washington and enjoys exploring the PNW in her free time.
Claire Palmer is a Solutions Architect at Amazon Web Services. She is on the Global Account Development team, supporting healthcare and life sciences customers. Claire has a passion for driving innovation initiatives and developing solutions that are both secure and scalable. She is based out of Seattle, Washington and enjoys exploring the PNW in her free time.